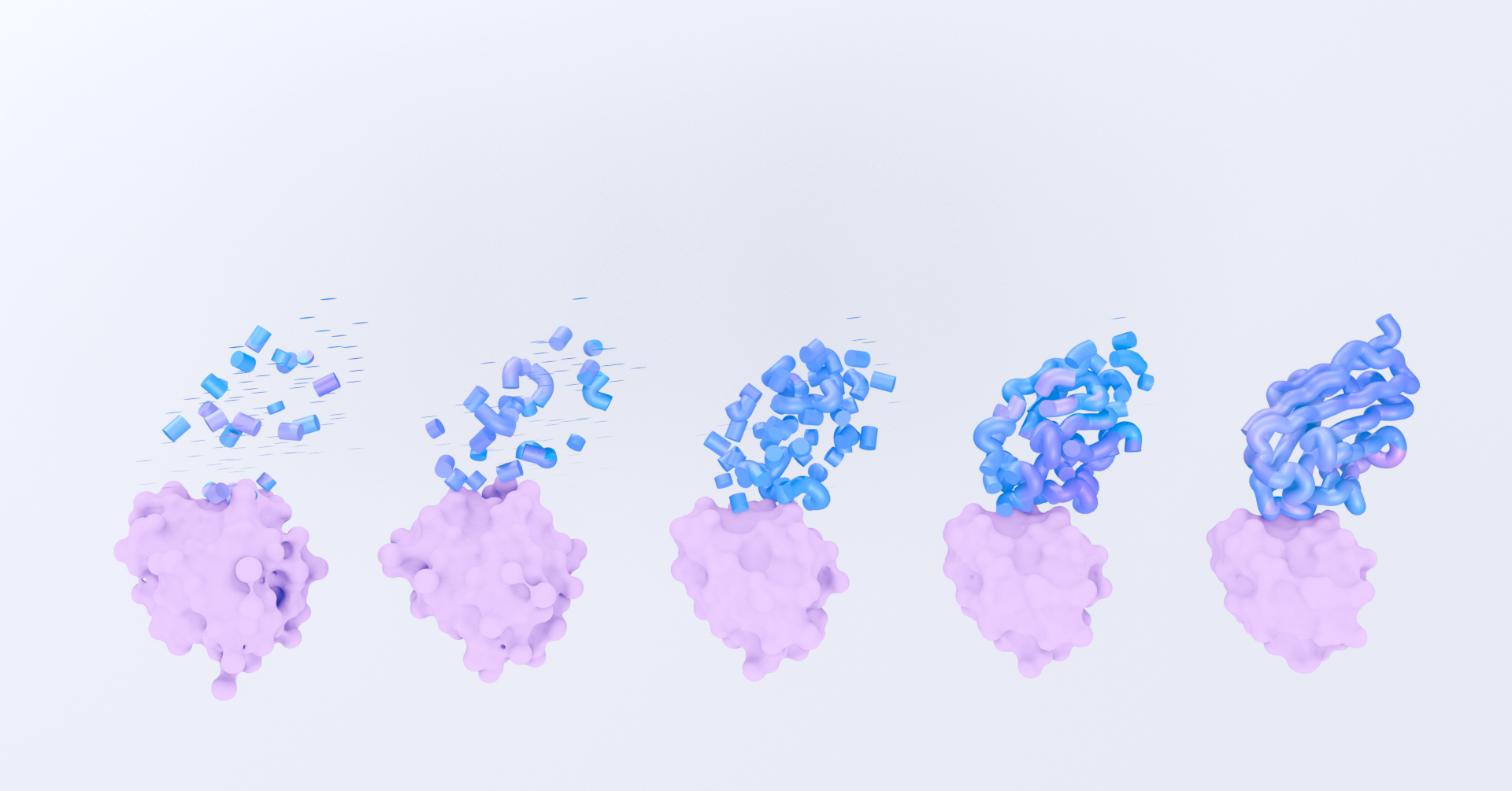
As part of our mission to engineer new and useful proteins with unprecedented control and accuracy, Diffuse is opening public access to our AI models for the first time.
Today we're releasing DSG2-mini, our newest AI protein design model. DSG2-mini is a compute-efficient version of our most advanced foundation model, DSG2. We’re making this model available in DiffuseSandbox, our new app for researchers to explore protein binder design against targets of interest.
We currently support nanobody design on DiffuseSandbox, with support for scFvs and minibinders coming soon.
Click here to access DiffuseSandbox.
Our previous model, DSG1, demonstrated breakthrough capabilities in protein design tasks. DSG2-mini exceeds DSG1 and open-source models RFDiffusion1 and RFAntibody2 significantly for protein design tasks. To learn more, see our benchmarking data here.
DiffuseSandbox makes it easy for scientists to test out advanced AI protein design capabilities without requiring a computational background. We hope that an open platform to investigate and interrogate the current capabilities of our models will accelerate innovation across the AI x Bio community.
To get started:
If it’s of interest to design large libraries of AI-designed proteins, please get in touch. We're look forward to your feedback!
DiffuseSandbox gives you a way to explore our newest protein design model DSG2-mini in an interactive way. We’re planning to deploy new binder design capabilities, including minibinder and scFv design, as well as enabling API access in the near future.
If what we’re working on interests you, we encourage you to try DiffuseSandbox, join our team, or get in touch to explore partnership opportunities and more extensive use of our pipeline.
— The Diffuse team
1 RFDiffusion
2 RFDiffusion for antibodies
Here, we report benchmarking data for DSG2-mini the task of nanobody (single domain antibody) design.
To benchmark our computational nanobody design capabilities, we used protein crystal structures from our clustered internal test set. Models are prompted to generate thousands of designs of nanobodies to bind to a target. Models are evaluated on their ability to generate designs with realistic interfaces, that are well-packed, and that are predicted to bind in laboratory experiments.
We find that DSG2-mini outperforms DSG1, RFdiffusion1, and RFantibody2 on key metrics relating to protein monomer quality and protein-protein interface design, and it approaches the natural distributions of these metrics for ground-truth protein 3D structures.
.png)



We are eager to connect with you.